Community lab update #4: April 2025
A call for a Monstera deliciosa reference genome and structured plant tissue culture data
👋 Hi friends,
Did you know that the most popular houseplant—our beloved Monstera deliciosa—actually produces edible fruit? In fact, its specific epithet deliciosa is a reference to its large fruit that tastes like a mixture of pineapple, banana, and mango, although it looks more like a green ear of corn.
So why doesn’t your living room Monstera ever bear fruit? It’s all about the environment. Indoors, these tropical plants rarely produce their tasty fruit. Travel to their native rainforests in Mexico and South America, or even to tropical Florida, and you’ll find Monsteras climbing high with enormous leaves and, in the right conditions, crowned with their corn-shaped fruit. If you do take a bite, be careful—only eat it when it's ripe. Unripe fruit contains calcium oxalate crystals that can cause a burning sensation in your throat and mouth.



Since we’re on the topic, let me share one more thing before we dive into April’s update. The world has fallen in love with variegated Monstera deliciosa cultivars. Remember when collectors (myself included) lost it over Costa Farms repeatedly delaying the release of their M. deliciosa ‘Thai Constellation’ three years ago? To calm the frenzy, they released a small batch online and it sold out in minutes despite the $600 price tag. And that’s nothing compared to the $17,500 someone paid for the M. deliciosa ‘Bulbasaur’ cultivar. Don’t worry, there’s still one left if you’ve got some spare change. But wild as that is, the visual trait behind all this hype—variegation—can make a scientist’s eyes light up just as much as a houseplant collector’s.
Variegation often reflects underlying changes in gene activity, like mutations in the genome, disruptions in pigment biosynthesis, or differences in protein expression. These differences can affect how chloroplasts develop and function, which in turn influences whether variegation is stable, unstable, or in rarer cases, heritable. In M. deliciosa ‘Thai Constellation’, variegation is relatively stable, while in ‘Mint’ and ‘Albo’ cultivars, it's unstable because the mutations affect only some cell layers, a condition known as chimerism. You may have seen this in action if your ‘Mint’ or ‘Albo’ leaves have ever reverted to all green, or worse, all white. But to truly understand the molecular basis behind these differences, we’re missing a key piece: a Monstera deliciosa reference genome.



Fortunately, the chloroplast genome (plastome) of Monstera deliciosa was sequenced about two years ago, but we still lack its full nuclear genome. There are a few reasons for this. First, while plastome mutations have been linked to albino phenotypes in some plants, it's unclear if that's true for Monstera. To find out, we’d need to sequence the plastomes from green and white tissue in cultivars like ‘Thai Constellation’ or ‘Albo’—but we may find no differences at all, suggesting the cause lies in the nuclear genome. Second, the nuclear genome is large—about 4.9 Gb and nearly 30 times the size of the plastome—and likely full of repetitive elements, making genome assembly difficult. Still, the plastome offers a useful starting point. Sequencing the full genome could uncover the genetic basis of variegation and even enable gene knockouts to reduce calcium oxalate in the fruit. There’s a lot to discover. We could literally enjoy the fruit of our labor.
To make this happen, I need your help. Sequencing the M. deliciosa genome isn’t trivial, so I’m in conversation with a few plant genomics labs that have the expertise and infrastructure to take this on. But to move forward, we need to show that the plant community wants this. Whether you're a plant synthetic biologist interested in gene editing for variegation, or simply a curious plant lover dreaming of oxalate-free fruit, your voice matters. Please like, comment, or share this post to show your support! From sample prep to sequencing and genome assembly, we’ll handle the entire process and keep you updated every step of the way. With a reference genome in hand, we can begin to explore the genetic basis of variegation, reduce calcium oxalate in the fruit, and unlock even more possibilities at the community plant biology lab.
Finally, and I promise I’m almost done, let’s talk about why knocking out genes involved in calcium oxalate production in M. deliciosa fruit is such a powerful idea. The same workflow—genome sequencing and assembly, gene identification, and editing with CRISPR-Cas systems—can also be used to rapidly domesticate orphan crops in ways that weren’t historically possible. If you attended last night’s CRISPR talk with Evan Groover, this probably sounds familiar. It’s the very approach scientists across Africa are using to make understudied crops more edible, nutritious, and farmer-friendly. That’s why I’m so passionate about using Monstera deliciosa as a lab model for a community plant biology lab. Its popularity and accessibility make it an ideal entry point for teaching advanced techniques with real-world impact—empowering more people to participate in the future of plant science.



P.S. I’m also teaching a two-day plant tissue culture workshop at Genspace in Brooklyn on June 7th and 8th. If you’re ready to go deeper down the Monstera and tissue culture rabbit hole, join me!
P.P.S. I recently had the pleasure of attending a private screening of the Observer, a film that explores the magic of observation in science and the broader human experience. It reminded me of the child-like curiosity surrounding science that seems to have gotten lost as the “institution of science” emerged. So, I’m working with the Science Communication Lab to arrange one or two screenings of the film with a group discussion/brainstorm to follow. Stay tuned for more details!
Now, as promised, let’s dive into April’s lab update.
Metrics
Burn: $294.33
Initiated cultures: 60
Contamination rate: 20/60, 33.3%
Time spent per culture: ~22 minutes (23.5 hours total for 60 new cultures and 3 subcultures)
What happened in April
I submitted my application to the Astera residency and received notice that my O’Shaughnessy fellowship is under review. 🤞
I attended the Conserving Exceptional Plants Symposium on April 8-9, 2025. This led to a great conversation with Devon Gordon, the Manager of the Hawai’i Micropropagation Lab, where he oversees efforts to conserve over 50,000 plants via tissue culture.
In an effort to make plant cell and tissue culture interoperable, I began circulating a request for information (RFI) to explore how to design a standard data framework, identify key data types for machine learning, and uncover potential predictive use cases. (Thank you to all the collaborators who have gone back-and-forth with me on this over the past month!)
Why does this matter? Plant cell and tissue culture underpins efforts in genetic engineering, plant conservation, metabolite production, and commercial horticulture. And, data is being generated across commercial labs, botanical gardens, academic labs, and even by a growing community of plant tissue culture hobbyists. Yet, current protocol development is slow and guesswork-heavy and researchers often repurpose protocols from related species or test endless combinations without a shared framework. Data interoperability could enable machine learning models to predict ideal culture conditions across species—accelerating propagation of endangered plants or understudied species with unique use cases.
The results from the Monstera sterilization tests are in! For those of you who are new here, my plant tissue culture work focuses on rare houseplants and often I only have access to small amounts of “starting” tissue, sometimes no more than a single leaf or in the case of Monstera species, a few nodes. As a result, it’s important to hone in on a sterilization process so that the odds of successful cloning are as high as possible. After running 264 experiments to test different sterilizing agents—sodium hypochlorite (NaOCl) and sodium dichloroisocyanurate (NaDCC)—at varying concentrations and exposure times, it looks like the verdict is still out. Let’s review the results.
As we increase the concentration of each sterilizing agent, we would expect to see a lower contamination rate. The same is true when we increase the amount of time the nodes are agitated in our sterilization solutions. Intuitively this makes sense, as stronger and longer sterilization treatments should kill more microbes. Yet, at some point, the concentration will be too high, or the time spent in the solution will be too long, and we’ll get sterilized nodes with severely damaged tissue.
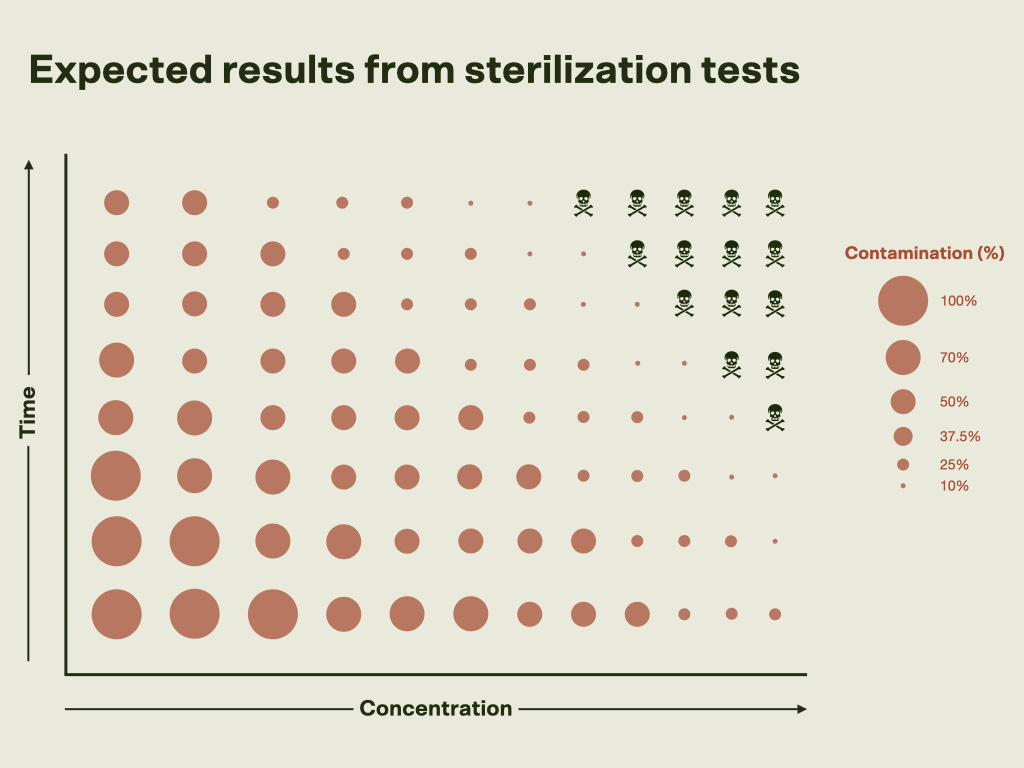
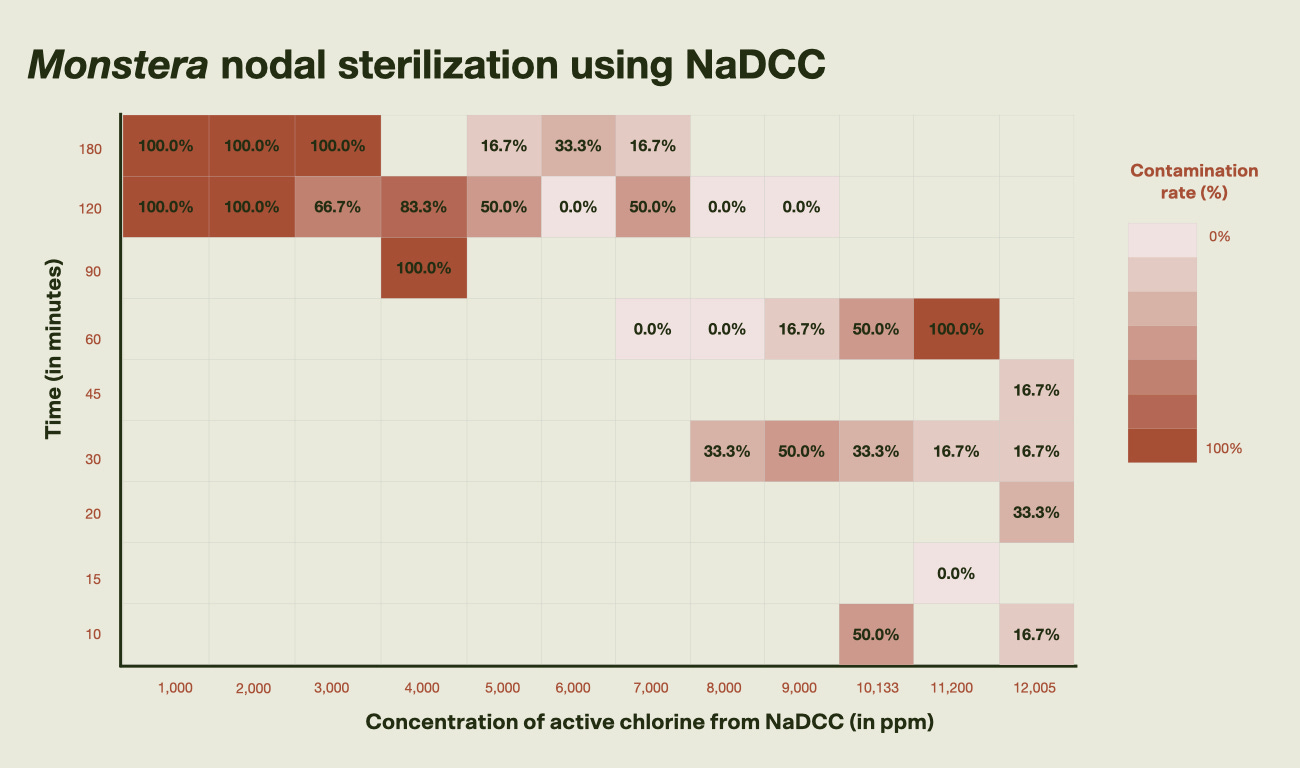
Figure 1: Hypothesized results from sterilization experiments. As time and concentration of the sterilizing agent increases, we should see lower contamination rates. The goal is to find the condition that requires the lowest concentration for the shortest amount of time so that the explant sterilization process is quick, yet effective. Focusing on the NaDCC data, we see the expected trend. Contamination generally decreases as concentration and exposure time increase, though a few outliers exist. However, even at the highest concentrations tested—10,133 to 12,005 ppm active chlorine—contamination wasn’t consistently minimized. Nodes sterilized at 11,200 and 12,005 ppm showed a slight increase in browning, likely due to tissue oxidation (a natural stress response where plants release protective compounds when damaged). Browning was reduced when 50 mg/L of ascorbic acid, an antioxidant, was added to the media during the 12,005 ppm tests. Regardless of the presence of ascorbic acid, shoot growth was observed within three weeks of sterilization.
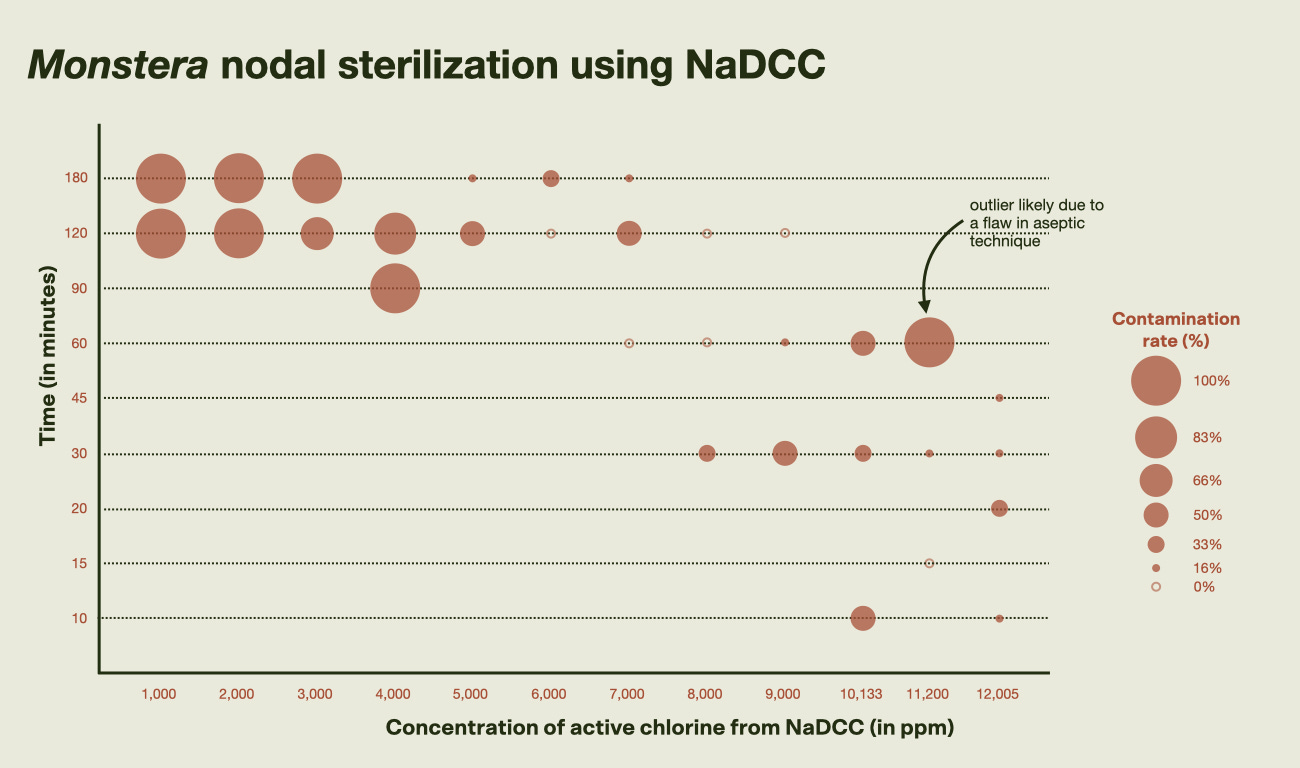
Figures 2 and 3: Monstera deliciosa nodal sterilization outcomes using varying concentrations of sodium dichloroisocyanurate (NaDCC) and exposure times. Six replicates were tested per condition, with no more than three explants placed in the same 50 mL tube containing the sterilization solution. Lower contamination rates were generally observed at higher concentrations and shorter exposure times. One outlier with unexpectedly high contamination at 11,200 ppm for 60 minutes is likely due to a flaw in aseptic technique. Note that the y-axis (exposure time) is not evenly scaled across all intervals. Since tissue damage appears minimal—given consistent shoot growth—and contamination rates haven't stabilized between 10-16%, the sweet spot may lie just beyond the tested range. I’ll test exposures of 13,000 to 14,000 ppm for 10–30 minutes during this May’s lab work.
To kick off climate week, I co-taught another plant tissue culture workshop at Counter Culture Labs in Oakland on April 20, 2025.
What’s happening in May
I spoke about how a community plant biology lab can be financially supported by the commercial sale of rare houseplants at the BioPunk (Un)conference on Saturday, May 3rd at 11:30 AM. (Big thanks to Elliot Roth for the invitation!)
Evan Groover and I hosted a plant swap and CRISPR discussion at Kinfolx on Thursday, May 15th from 6-8 PM.
I’ll finish a taxonomic distribution analysis I’m working on to show which parts of the plant kingdom are overstudied—likely commercial, medicinal, and model species—and which are understudied. This will help guide the development of testing a standardized plant cell and tissue culture data and metadata framework.
I have a ton of subcultures and experiments to catch up on so I need to prioritize those before applying to the Emergent Ventures grant and initiating new species.
How you can help
Like, comment, and/or share this post to help me collaborate with scientists to sequence the Monstera deliciosa genome! 🪴
If you’re familiar with plant cell and tissue culture, I’d love to get your input. See the RFI for specific questions of interest, but I especially want to know why not to pursue this effort. I have a few ideas myself—the plant conservation and genetic engineering fields are moving away from plant tissue culture, embracing cryopreservation and pursuing pollen, seed, and meristem transformation. What other scenarios would make this effort obsolete?
Please keep the encouragement coming! I receive so many texts, calls, and emails from you all whether it's about potential funding opportunities, scientific feedback, donating lab equipment, or just letting me know you’re rooting for this lab. Although it’s hard for me to respond to everything quickly (eventually I do respond), it means the world to me. Thank you. 🙏
Feel free to share this with any friends who might be interested. And if you received this email from someone else, you can subscribe directly to get the next one. 🌵





I'd love to see what new varieties people come up with if they had a M. Deliciosa reference genome! Also the Observer looks beautiful, hope you screen it!
It’s so interesting how popular variegated Monstera are when it’s simply the result of genetic mutation! What challenges do you anticipate with sequencing such a large genome?